Últimos assuntos
Tópicos mais visitados
Tópicos mais ativos

ENCODE: The human encyclopaedia
Página 1 de 1
06092012

 ENCODE: The human encyclopaedia
ENCODE: The human encyclopaedia

ENCODE: enterrou de vez a pseudociência do DNA "lixo" - Design Inteligente vindicado!
quinta-feira, setembro 06, 2012
NATURE | NEWS FEATURE
ENCODE: The human encyclopaedia
First they sequenced it. Now they have surveyed its hinterlands. But no one knows how much more information the human genome holds, or when to stop looking for it.
Brendan Maher
05 September 2012
Ewan Birney would like to create a printout of all the genomic data that he and his collaborators have been collecting for the past five years as part of ENCODE, the Encyclopedia of DNA Elements. Finding a place to put it would be a challenge, however. Even if it contained 1,000 base pairs per square centimetre, the printout would stretch 16 metres high and at least 30 kilometres long.
ENCODE was designed to pick up where the Human Genome Project left off. Although that massive effort revealed the blueprint of human biology, it quickly became clear that the instruction manual for reading the blueprint was sketchy at best. Researchers could identify in its 3 billion letters many of the regions that code for proteins, but those make up little more than 1% of the genome, contained in around 20,000 genes — a few familiar objects in an otherwise stark and unrecognizable landscape. Many biologists suspected that the information responsible for the wondrous complexity of humans lay somewhere in the ‘deserts’ between the genes. ENCODE, which started in 2003, is a massive data-collection effort designed to populate this terrain. The aim is to catalogue the ‘functional’ DNA sequences that lurk there, learn when and in which cells they are active and trace their effects on how the genome is packaged, regulated and read.
After an initial pilot phase, ENCODE scientists started applying their methods to the entire genome in 2007. Now that phase has come to a close, signalled by the publication of 30 papers, in Nature, Genome Research and Genome Biology. The consortium has assigned some sort of function to roughly 80% of the genome, including more than 70,000 ‘promoter’ regions — the sites, just upstream of genes, where proteins bind to control gene expression — and nearly 400,000 ‘enhancer’ regions that regulate expression of distant genes (see page 57)1. But the job is far from done, says Birney, a computational biologist at the European Molecular Biology Laboratory’s European Bioinformatics Institute in Hinxton, UK, who coordinated the data analysis for ENCODE. He says that some of the mapping efforts are about halfway to completion, and that deeper characterization of everything the genome is doing is probably only 10% finished. A third phase, now getting under way, will fill out the human instruction manual and provide much more detail.
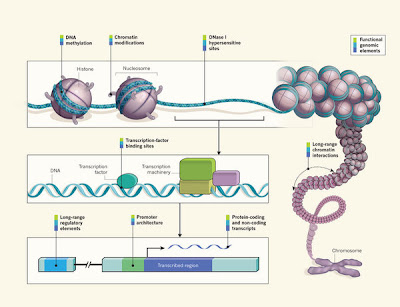
Many who have dipped a cup into the vast stream of data are excited by the prospect. ENCODE has already illuminated some of the genome’s dark corners, creating opportunities to understand how genetic variations affect human traits and diseases. Exploring the myriad regulatory elements revealed by the project and comparing their sequences with those from other mammals promises to reshape scientists’ understanding of how humans evolved.
Yet some researchers wonder at what point enough will be enough. “I don’t see the runaway train stopping soon,” says Chris Ponting, a computational biologist at the University of Oxford, UK. Although Ponting is supportive of the project’s goals, he does question whether some aspects of ENCODE will provide a return on the investment, which is estimated to have exceeded US$185 million. But Job Dekker, an ENCODE group leader at the University of Massachusetts Medical School in Worcester, says that realizing ENCODE’s potential will require some patience. “It sometimes takes you a long time to know how much can you learn from any given data set,” he says.
Even before the human genome sequence was finished2, the National Human Genome Research Institute (NHGRI), the main US funder of genomic science, was arguing for a systematic approach to identify functional pieces of DNA. In 2003, it invited biologists to propose pilot projects that would accrue such information on just 1% of the genome, and help to determine which experimental techniques were likely to work best on the whole thing.
The pilot projects transformed biologists’ view of the genome. Even though only a small amount of DNA manufactures protein-coding messenger RNA,for example, the researchers found that much of the genome is ‘transcribed’ into non-coding RNA molecules, some of which are now known to be important regulators of gene expression. And although many geneticists had thought that the functional elements would be those that are most conserved across species, they actually found that many important regulatory sequences have evolved rapidly. The consortium published its results3 in 2007, shortly after the NHGRI had issued a second round of requests, this time asking would-be participants to extend their work to the entire genome. This ‘scale-up’ phase started just as next-generation sequencing machines were taking off, making data acquisition much faster and cheaper. “We produced, I think, five times the data we said we were going to produce without any change in cost,” says John Stamatoyannopoulos, an ENCODE group leader at the University of Washington in Seattle.
...
Read more here/Leia mais aqui: Nature
NATURE ENCODE
+++++
NOTA DESTE BLOGGER:
A vindicação impressionante de uma previsão dos teóricos do Design Inteligente de que o genoma, longe de ser cheio de "lixo" sem função, iria se revelar como tendo funcionalidade em grande escala:
No livro The Myth of Junk DNA, Jonathan Wells escreveu:
Far from consisting mainly of junk that provides evidence against intelligent design, our genome is increasingly revealing itself to be a multidimensional, integrated system in which non-protein-coding DNA performs a wide variety of functions. If anything, it provides evidence for intelligent design. Even apart from possible implications for intelligent design, however, the demise of the myth of junk DNA promises to stimulate more research into the mysteries of the genome. These are exciting times for scientists willing to follow the evidence wherever it leads.
(Jonathan Wells, The Myth of Junk DNA, pp. 9-10 (Discovery Institute Press, 2011).)
Por muitos anos, a defesa do DNA"lixo" pelos evolucionistas - atrelados caninamente ao paradigma neodarwinista - impediu que a ciência fizesse grandes descobertas sobre o genoma humano! Então, cara-pálidas da Nomenklatura científica, qual teoria ou teóricos realmente impede o avanço da ciência? Galera dos meninos e meninas de Darwin, nota do tio Neddy: não pode mais usar o DNA "lixo" como argumento contra a teoria do Design Inteligente, visse?
Fui, nem sei por que, rindo, mas rachando de rir da cara de alguns mandarins da Nomenklatura científica tupiniquim que fizeram carreira e retórica inflamada em cima do DNA "lixo" como sendo um fato científico contra a teoria do Design Inteligente. Uma hora dessas eles devem ter enfiado o rabo entre as pernas...

Eduardo- Mensagens : 5997
Idade : 54
Inscrição : 08/05/2010
Permissões neste sub-fórum
Não podes responder a tópicos
 Início
Início








 Seja fã Forumeiros
Seja fã Forumeiros
» Acordem adventistas...
» O que Vestir Para Ir à Igreja?
» Ir para o céu?
» Chat do Forum
» TV Novo Tempo...
» Lutas de MMA são usadas como estratégia por Igreja Evangélica para atrair mais fiéis
» Lew Wallace, autor do célebre livro «Ben-Hur», converteu-se quando o escrevia
» Ex-pastor evangélico é batizado no Pará
» Citações de Ellen White sobre a Vida em Outros Planetas Não Caídos em Pecado
» Viagem ao Sobrenatural - Roger Morneau
» As aparições de Jesus após sua morte não poderiam ter sido alucinações?